In silico Transcriptome (iTranscriptome)
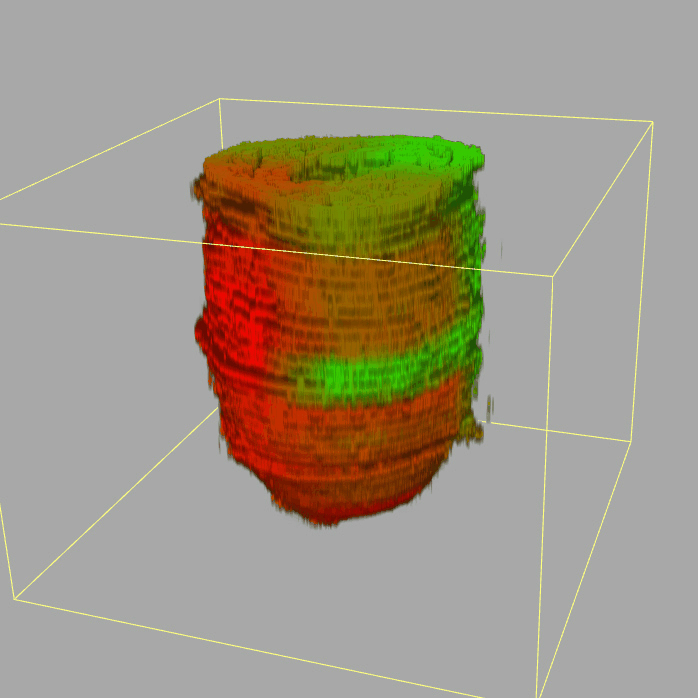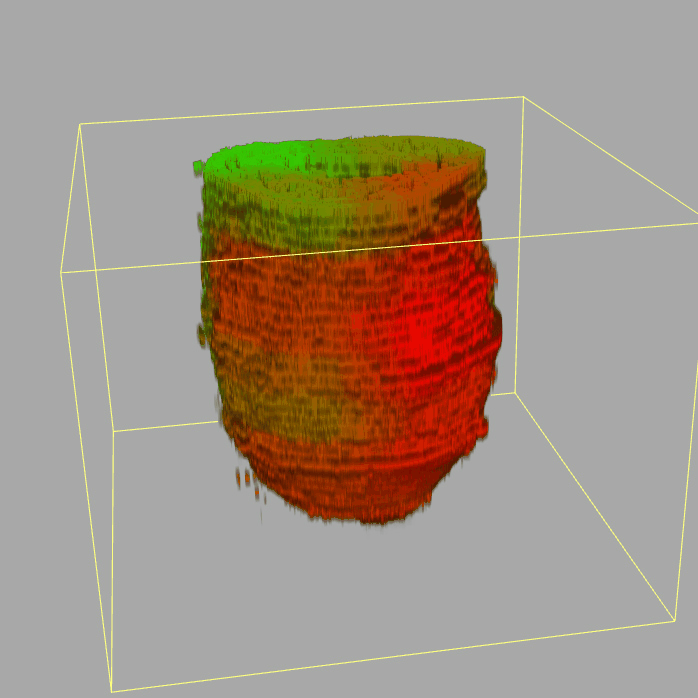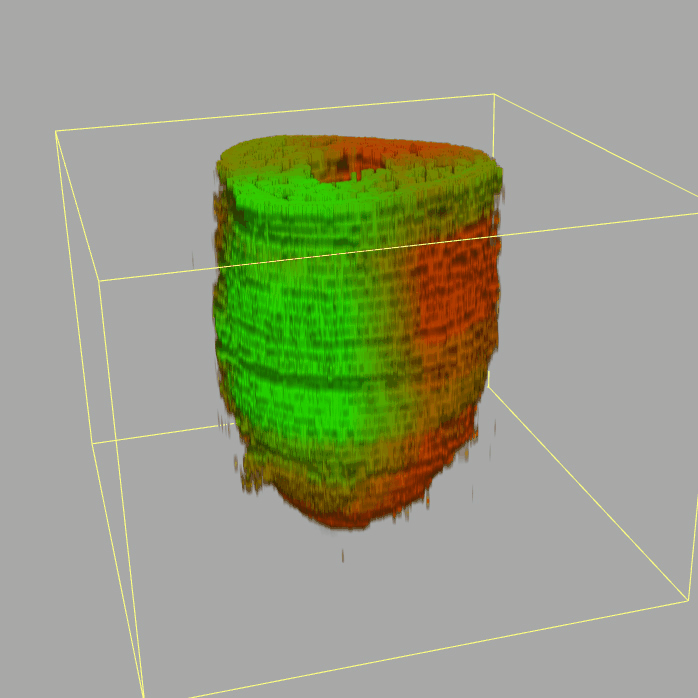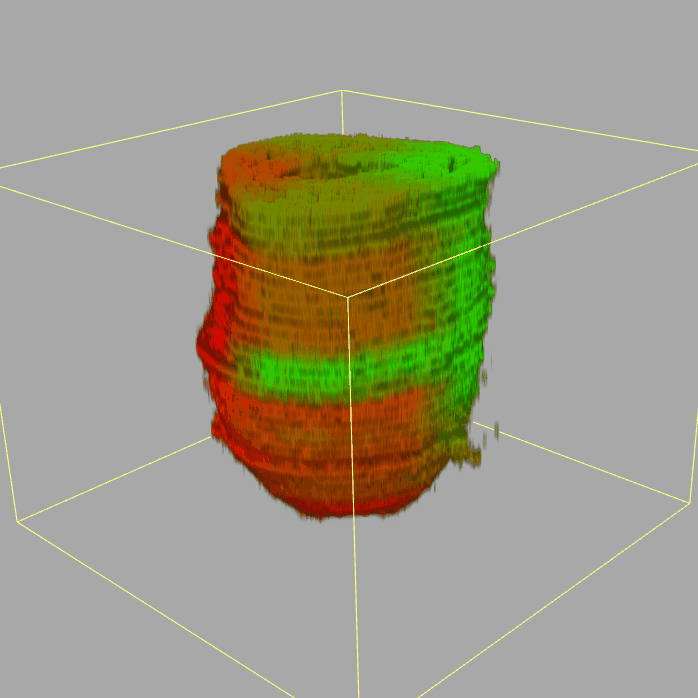
Geo-seq: 3D Transcriptome of the mouse embryo at gastrulation
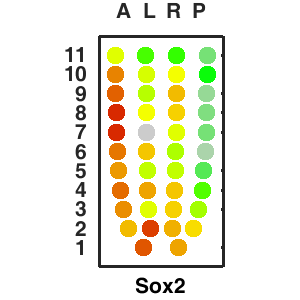
| Top 10 gene with the highest corrlation to : |
To download all the related data, please click
here. |
Key features |
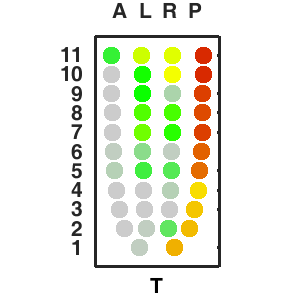
| - High resolution |
- More sensitive |
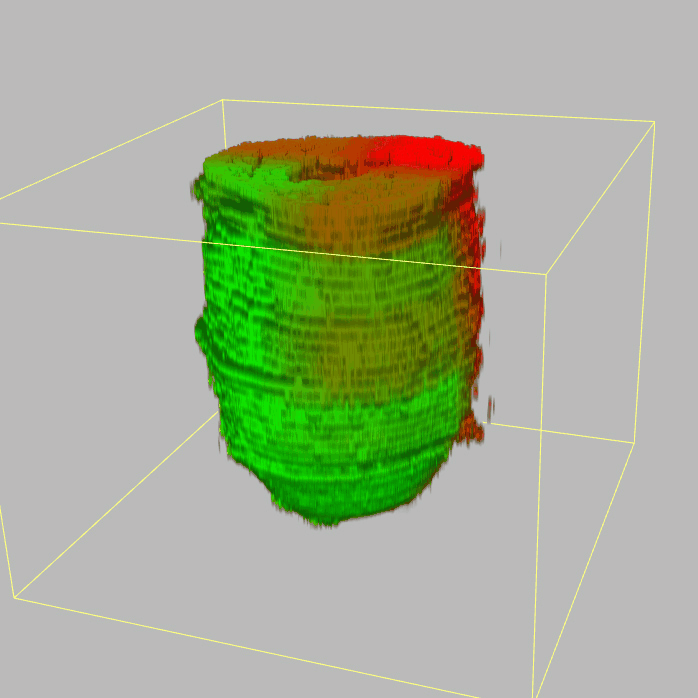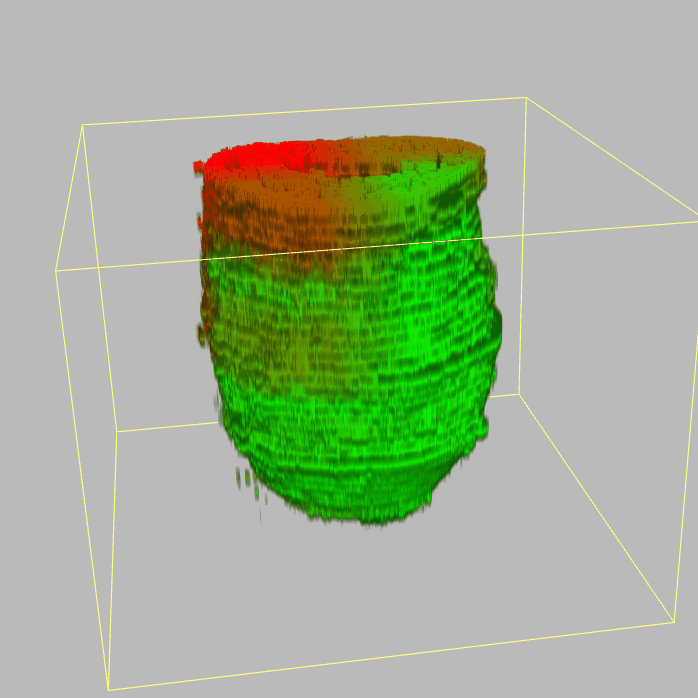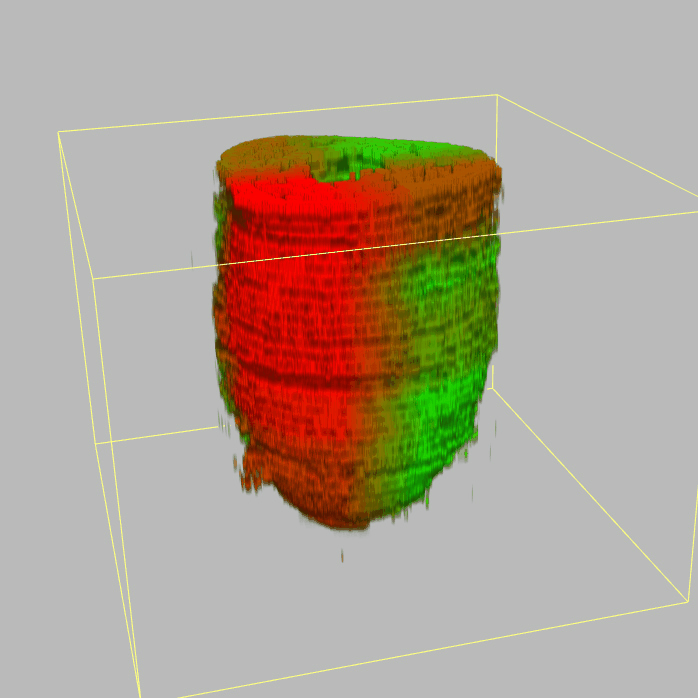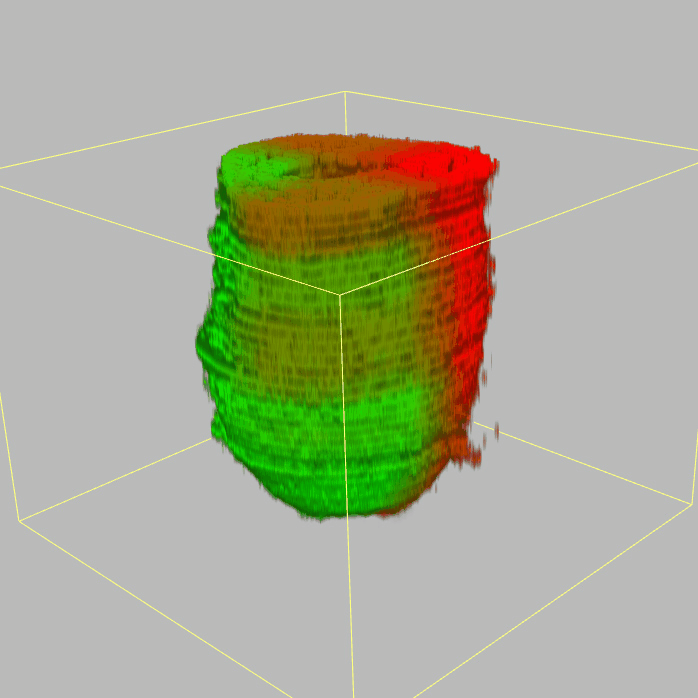
| - 2D & 3D visualization |
- Searchable web service |
| |
|
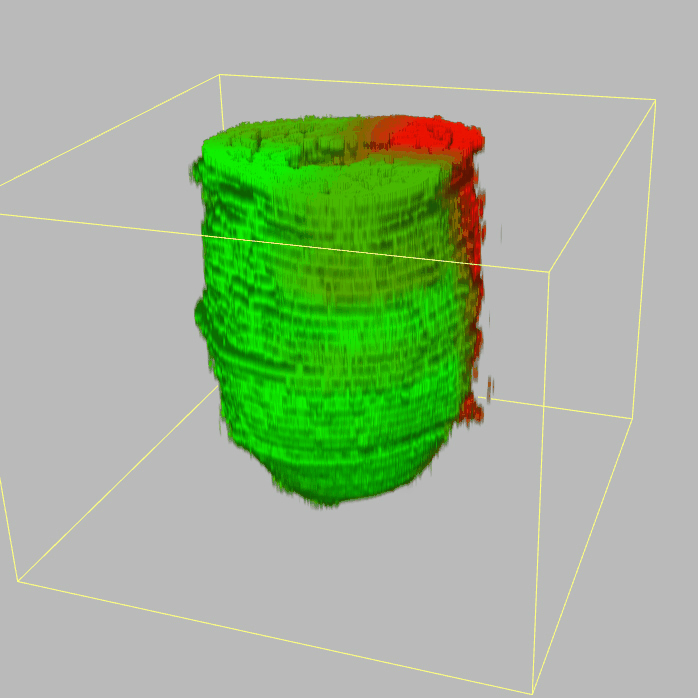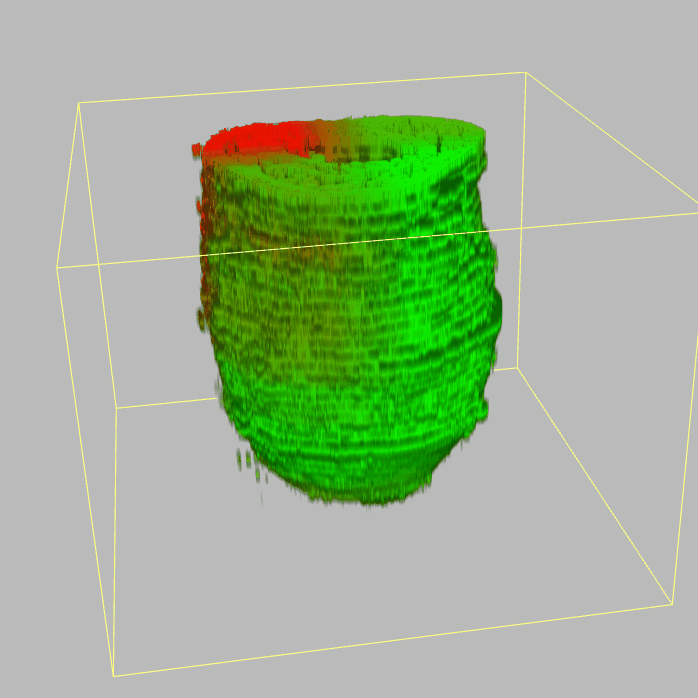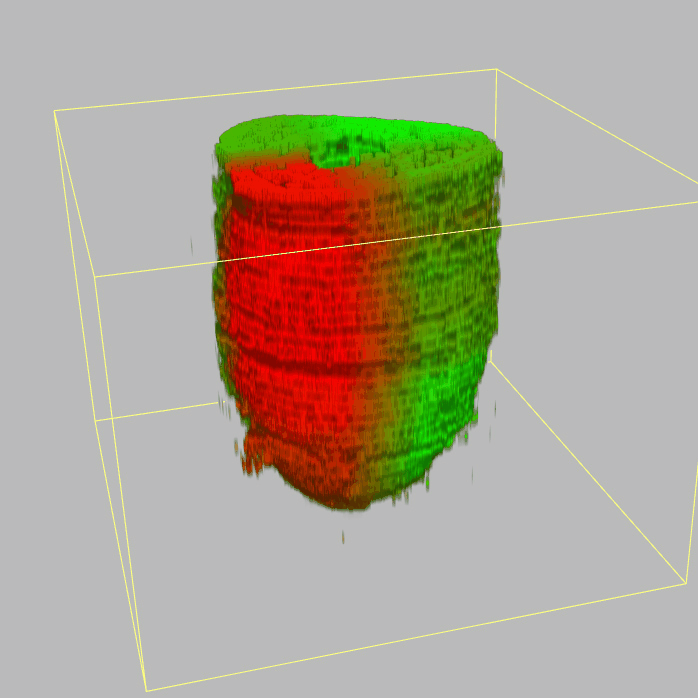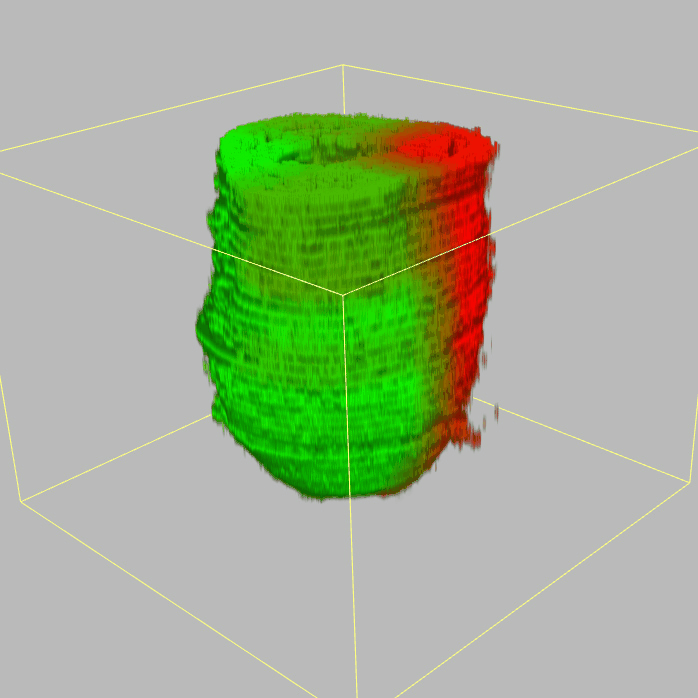
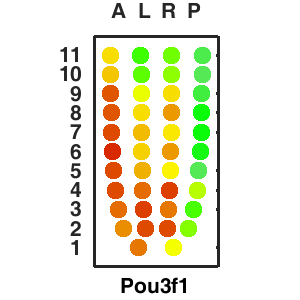
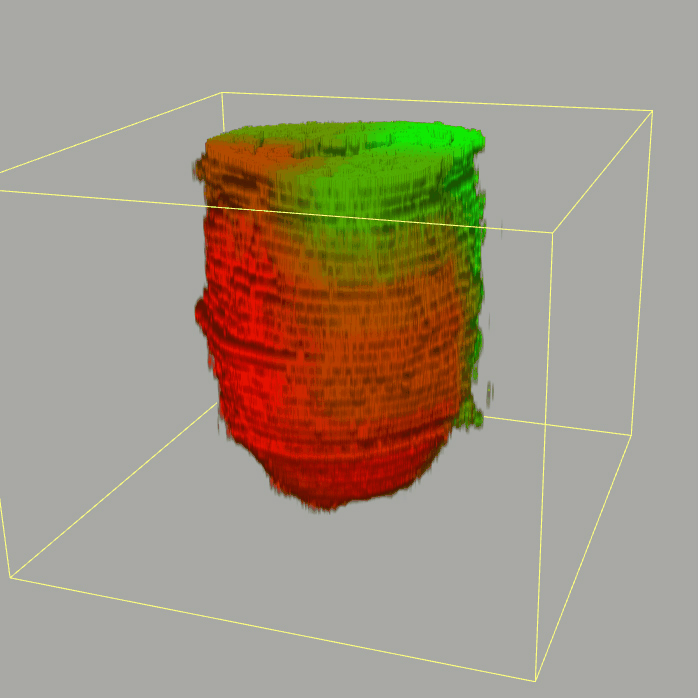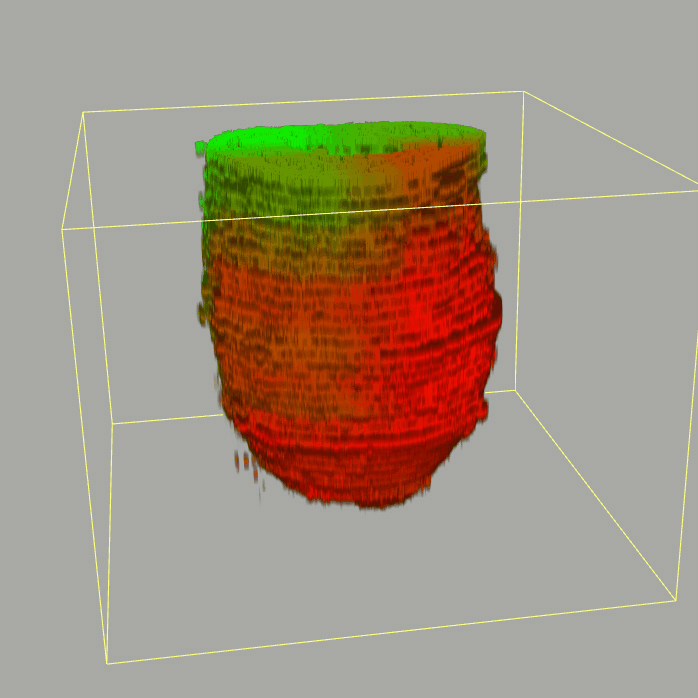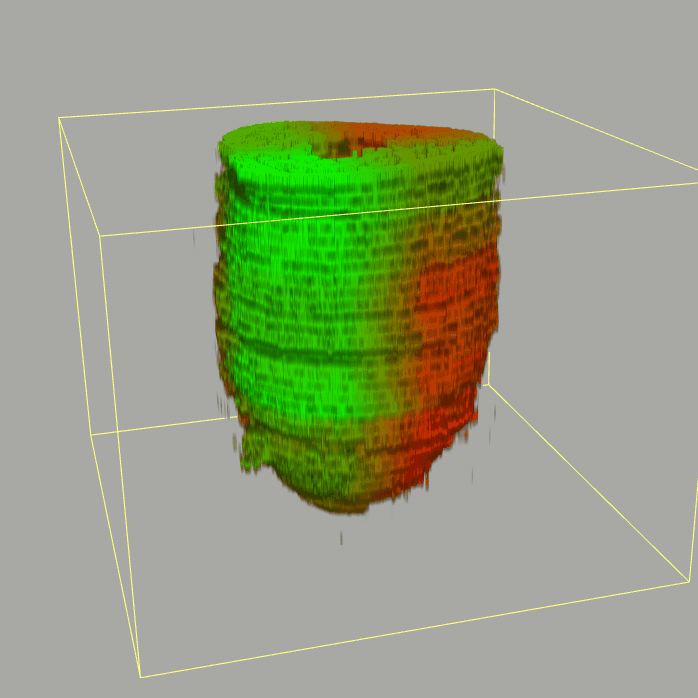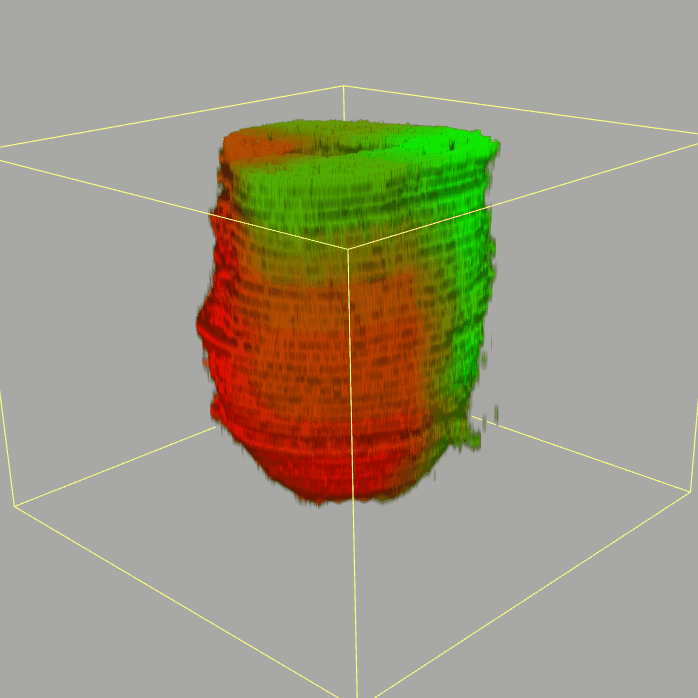
Query and display the expression pattern of a query gene in either a corn plot or a 3D embryo model
Search for genes that share similar expression patterns of a query gene.
Search for genes using a query pattern (Choose one of 5 predefined patterns or input interested pattern).
Calculate the enrichment score for a query gene list based on ranked gene expression of each sample.